Why ATTR is Ideal for In Vivo Editing
Transthyretin amyloidosis (ATTR) is a systemic proteinopathy driven by TTR aggregate deposition, a tetrameric protein mainly produced in the liver. Current treatments (stabilizers, siRNA/antisense) lower circulating TTR and slow progression but fail to correct the genetic defect and typically require repeated lifelong administration. CRISPR-Cas9 (e.g., NTLA-2001) has demonstrated durable TTR reduction in humans and is in phase 3, but DSB + NHEJ can generate In-Frame Mutations (IFM), a nontrivial issue for an intrinsically amyloidogenic target protein.
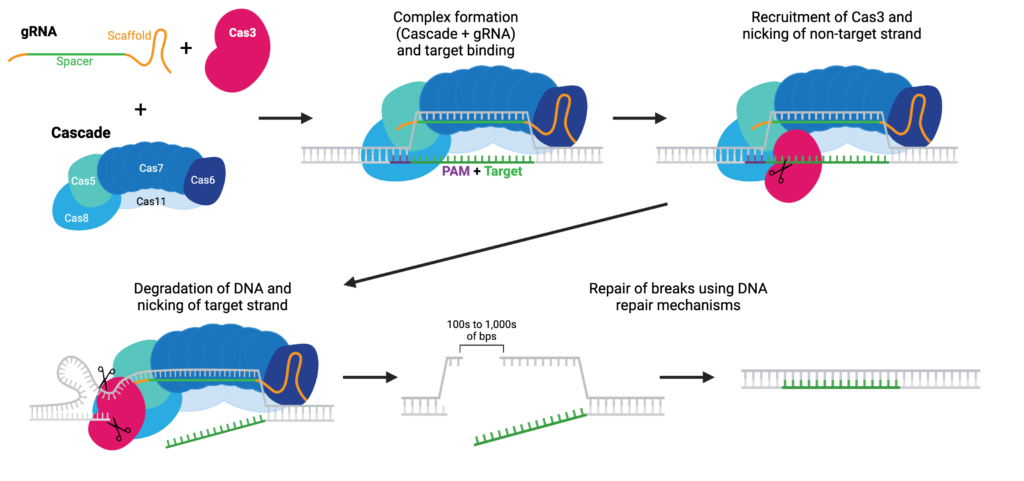
Cas3 in One Idea: Type I System That Erases, Doesn’t Patch
CRISPR-Cas3 belongs to class 1 (type I-E) CRISPR systems: target recognition is mediated by a multi-subunit Cascade complex (loaded with crRNA) that binds DNA, forms an R-loop, and recruits Cas3. Cas3 is a processive helicase-nuclease: once activated, it unwinds and degrades DNA unidirectionally, typically yielding long deletions (often upstream of PAM) rather than small indels from localized cuts. For “knock-out” indications (like TTR inactivation), this profile may minimize residual activity risk from short IFMs.
Key Results from the Team
- In vitro (Hepa1-6): Post-crRNA optimization, ~58.9 ± 0.5% editing at TTR/Ttr locus.
- Cas3 editing profile: Predominantly directional deletions up to ~75 kb in vitro.
- In vivo (mouse liver, LNP): Single injection yields 48.7 ± 1.1% hepatic editing and 80.1 ± 4.6% serum TTR reduction.
- In vivo deletion size: More confined under LNP conditions, max ~21 kb.
- Expression kinetics: Cas3 detectable in liver at 4–6 h, fades by 24 h, undetectable at 168 h.
- RNA transience: Cascade transcripts degraded in <1 week, supporting short, potentially safer activity.
Game-Changing Safety: Off-Targets and IFMs
Cas3 shows no reproducible off-target mutations (including post short-read WGS ~139× and long-read ~28×), unlike Cas9’s low-frequency events (e.g., POT1, POT129). The major distinction lies in editing products: in Ttr exon-humanized mice, Cas3 reduces human serum TTR by ~74.6 ± 0.6% (vs 94.9 ± 0.3% for Cas9/NTLA-2001), avoiding detectable IFMs. Conversely, Cas9/NTLA-2001 yields a mutant TTR peptide (IFM4, p.D39_40TdelinsA) via nanoLC-MS/MS, linked to enhanced amyloidogenicity in structural/biophysical assays.
Cardiac Benefits + LNP Tolerance: Cas3 “Bonuses”
Beyond TTR reduction, Cas3 lowers early cardiac pathology markers in exon-humanized models, including TTR deposition with macrophage accumulation (CD68 signal) via immunofluorescence. The study notes a transient cytokine response post-LNP injection (early peak, baseline by ~1 week), akin to empty LNPs, suggesting acceptable short-term tolerance.
Takeaways (and Watchpoints)
Key takeaway: Cas3 isn’t “better Cas9″—it’s a mechanistically distinct editor ideal for robust gene knockout while minimizing residual IFM risk. Watchpoints: Fine deletion size control, inter-context variability (plasmid vs mRNA-LNP), and long-term safety studies remain essential pre-clinical translation.
Corresponding author: Tomoji Mashimo – Journal: Nature Biotechnology (January 2026) – DOI: https://doi.org/10.1038/s41587-025-02949-6